
DNA is like a fingerprint. Each species’ DNA signature is distinctive. And it can be found on the scene long after the individual has gone.
For some time, scientists have gathered the DNA that organisms leave behind—in saliva, urine, feces, fur, and flakes of dry skin—to detect their presence and study them.
They’ve found this so-called environmental DNA, or eDNA, in water, soil, even snow.
But scientists wondered if they could gather it more easily and broadly by capturing it out of thin air.
To test this, they set up vacuum devices at zoos and sucked in the air, trapping all particles within it on very fine filters.
They then used a polymerase chain reaction—the same PCR methodology that’s become well known in Covid tests—to amplify DNA signatures of the particles.
Surprisingly, from DNA in the air alone, they could catalog the animals in the zoo, from huge giraffes to tiny guppies in ponds.
They also found eDNA of native animals, like squirrels and deer. Domesticated pets like cats and dogs. And of course, human visitors.
The technique was not perfect however, missing the presence of some of the biggest creatures.
But as it matures, gathering eDNA from the air has powerful potential to measure populations, track elusive animals, detect pathogens, even solve crimes.
Background
Synopsis: All living organisms shed DNA into their environment, and, when it can be detected, environmental DNA (eDNA) provides a noninvasive record of their presence. Scientists have long collected and studied eDNA from soil and water. Now, researchers have attempted to suck DNA out of the atmosphere—and it worked! eDNA techniques provide additional nondestructive ways for scientists to study terrestrial biodiversity, invasive species and pathogens.
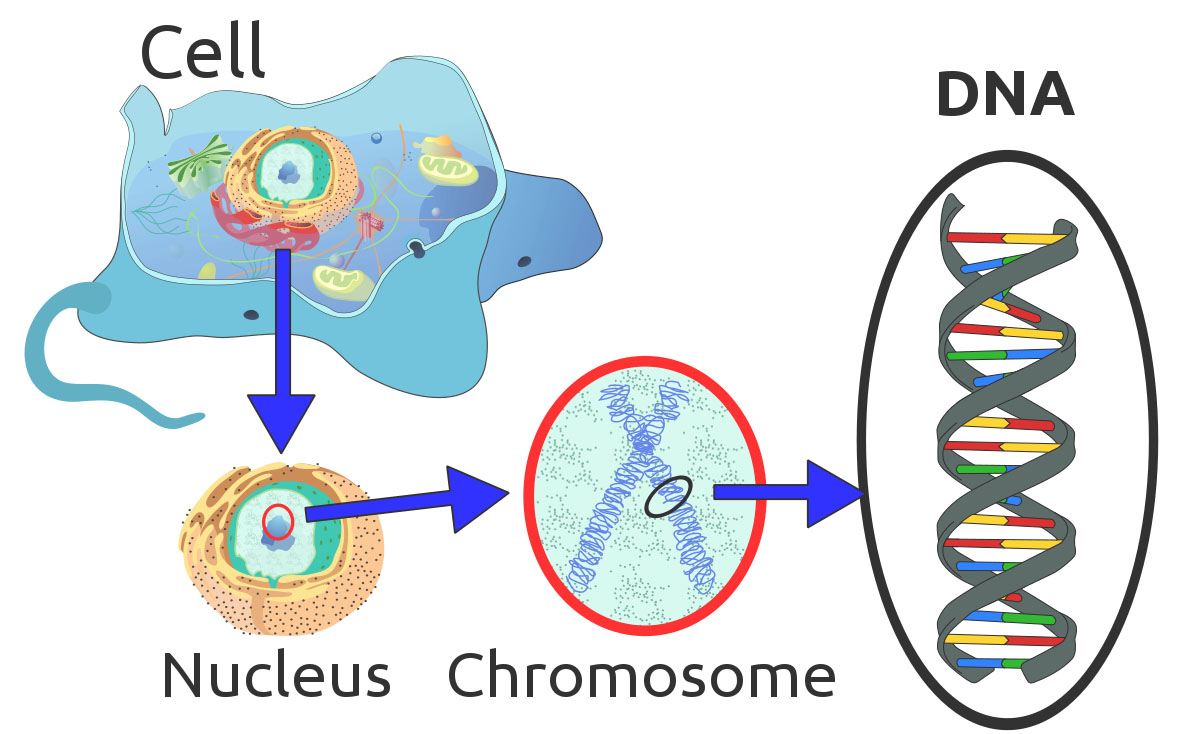
- Some researchers describe DNA as the universal language of living things.
- All known living cellular organisms that are capable of reproduction contain Deoxyribonucleic Acid (DNA) in every cell.
- DNA contains the instructions necessary for life and must be replicated for every living cell as well as being passed to offspring by parents.
- Viruses replicate using RNA but need to invade living cells to use their replication machinery. This dependence on other organisms to replicate is the reason they are not considered to be living organisms.
- All organisms shed DNA into their environment. This environmental DNA is known as eDNA.
- Every day, living organisms leave a wake of cells behind them in their saliva, urine, feces, hair or fur, scales and skin cells.
- Dead organisms leave DNA behind as they decay.
- Soil eDNA has been studied since the 1980s at botanical, anthropological and archaeological sites.
- Over the past decade researchers have adapted eDNA methods for nondestructive applications in aqueous systems.
- Fish migration and shark encroachment can be monitored without disturbing natural species movements with nets.
- Scientists can search for endangered species and keep track of reef health without impacting delicate ecosystems.
- Evidence of invasive species, as well as algal, bacterial, and fungal pathogens can be collected to detect early incursion before irreversible damage is done.
- A search for the Loch Ness Monster resulted in high concentrations of eel DNA.
- And, eDNA methods have been adapted to track eRNA from Covid-19 in sewage to predict viral outbreaks.
- To amplify eDNA signals in water, researchers employ a technique called polymerase chain reaction (PCR) to strengthen the faint signals.
- This is the same PCR methodology that has recently become famous as a key step in Covid-19 laboratory testing.
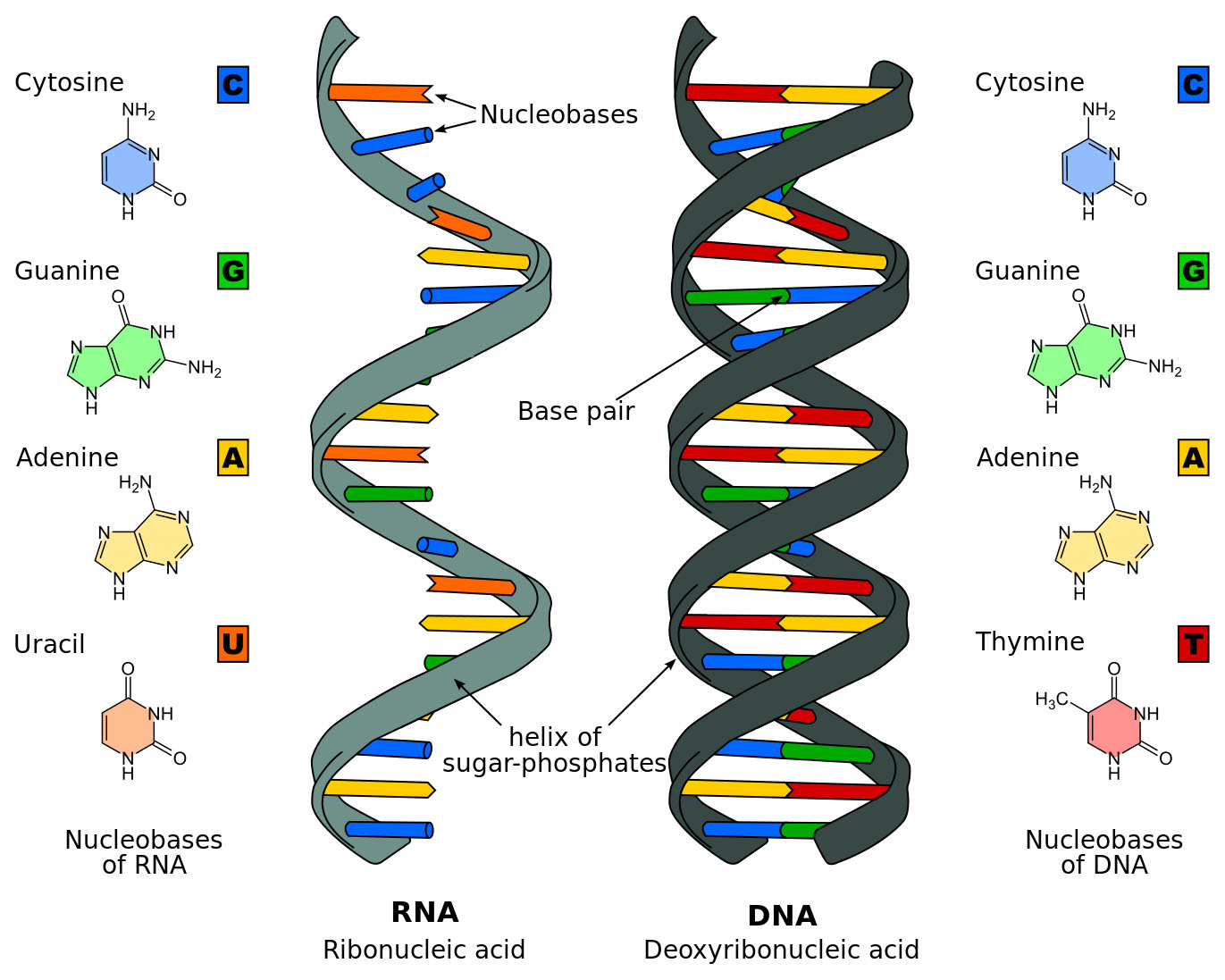
- Like a copy machine, the PCR process takes snippets of DNA and uses heat to split them apart right down the rungs of the double helix ladder.
- The rungs of the DNA ladder are made of pairs of the nitrogen bases: adenine (A), thymine (T), guanine (G) and cytosine (C). Adenine will only pair up to make rungs with thymine (A-T), and guanine will only latch onto cytosine (G-C).
- In other words, each sequence of bases (for example ACGTG) will only pair with an opposite, predictable sequence of bases (in this case it would have to be TGCAC).
- Then researchers cool the mixture to allow the segments to bind to matching sequences, creating two copies from the original fragment.
- As samples are heated and cooled repeatedly, they create up to a billion copies in a few hours, amplifying the DNA enough that it can be detected.
- Following successful applications in aqueous systems, the idea of collecting eDNA from the air was proposed and a few early experiments with plant DNA showed promise.
- Airborne eDNA collection is complicated by lower concentrations of particles dispersed in air.
- In 2020, medical researchers tested hospital room air and detected Covid-19 eRNA, a key clue in confirming that Covid-19 virus transmission is airborne.
- Recently, a few science teams set out to further test the eDNA analysis process in air by using vacuums and blowers to capture particles on filters.

- First, as proof of concept, a team of British scientists tested air in an enclosure that held a group of burrowing animals called naked mole rats. Amplification of the signal indicated the presence of DNA from both naked mole rats and their human caretakers.
- Then, the same team tried outdoor settings at the UK’s Hamerton Zoological Park, reasoning that the unique non-native species in a zoo would provide an unambiguous signal that would be distinct from the local background.
- They set up vacuum pumps in 20 locations both inside and outside of zoo buildings and let them run for 30 minutes, collecting a total of 72 samples.
- After PCR amplification, the team identified 25 species of mammals and birds, including 17 zoo animals and a few local animals like deer and hedgehogs. They were even able to detect eDNA from animal products used for feed.
- In some cases, they could sense animals nearly 1,000 ft (300 m) from their enclosures, and they could detect animals outside of their sealed indoor enclosures. But they missed the zoo’s maned wolves.
- Separately, but at the same time, a Danish team proposed a “crazy” idea for an innovation-driven high-risk grant program: vacuuming DNA from air.
- For the same reasons as the UK team, they selected three locations in the Copenhagen Zoo: the rainforest house, the okapi stable and a spot between animal enclosures.
- They collected 40 samples over periods of 30 minutes to 30 hours using three different types of samplers: a vacuum pump, a small 5V blower and a larger 24V blower.
- After PCR, they detected animal eDNA in every sample: a total of 49 species including 30 mammals, 13 birds, 1 reptile, 1 amphibian, and 4 fish—they were even able to detect a single guppy in a pond in the rainforest house. They also reported DNA from local fauna like squirrels, dogs and cats.
- Shorter distances and larger biomass made it more likely that specific animals would be detected.
- They were able to detect the very slow-moving sloths, but they missed the hippos.
- The two teams learned of each other’s studies and published both papers as complementary approaches confirming the same discovery.

- In a third study, Swedish scientists collected insect eDNA at three Swedish field sites.
- They identified 85 different species of moths, bees, beetles, flies, ants and wasps, as well as the DNA of some birds and mammals.
- Nondestructive eDNA data collection methods don’t require the death of the insects sampled like most traditional methods, and multiple species can be detected in a single sample.
- The eDNA survey missed some species that were detected by simultaneous traditional surveys, finding only 4 of the 48 species of moths collected on sticky traps, but it did detect 5 species that were not collected on the sticky traps.

- Airborne eDNA studies are in their infancy, so researchers are still fine-tuning methods and thinking about possible applications for this rapid and sensitive yet harmless approach to studying life forms in an area.
- Researchers need to understand how DNA begins its journey and how it moves through the air (aerosol scientists and meteorologists can help with this).
- Studying how far eDNA can travel in different environments and how fast it degenerates are other essential pieces of the puzzle.
- This method may be particularly helpful for stress-free monitoring of difficult-to-observe populations in burrows and caves.
- The method may indicate presence of a species in an environment but not how long the species was present or the size of a population.
- All three studies found evidence of many nearby animals, but none of them found every animal in proximity to the samplers. This means eDNA should be used as an overlapping complementary tool for biodiversity surveys.
- All the airborne sampling methods found human DNA, suggesting both the need to guard against contamination and the potential forensic uses.
- Since there are millions of species yet to describe on Earth—for instance it is estimated that we have described only about 20% of the 5.5 million insects on Earth—using eDNA to complement existing techniques will help with terrestrial biodiversity surveys of large scale.
- Once unusual DNA is located, the search for the source can begin.
- Scientists have now established that any environmental medium, including water, soil, rainfall, snow and air can harbor DNA.
- Will our future include permanently deployed eDNA vacuum systems for global terrestrial biomonitoring?

